

{{ packagename }} is developed at Pontificia Universidad Javeriana as part of the Epiverse-TRACE initiative.
{{ packagename }} is an R package that offers tools for estimating vaccine effectiveness (VE), using a series of epidemiological designs including cohort studies, test-negative case-control, and screening methods (Halloran, Longini, and Struchiner 2010). The current version of the package provides a set of features for preparing, visualizing, and managing cohort data, estimating vaccine effectiveness, and assessing the performance of the models. Test-negative design and screening method will be included in future versions.
Our stable versions are released on CRAN, and can be installed using:
install.packages("vaccineff", build_vignettes = TRUE)The current development version of {{ packagename }} can be
installed from GitHub using the
pak package.
if(!require("pak")) install.packages("pak")
pak::pak("{{ gh_repo }}")Or using the remotes package
if(!require("remotes")) install.packages("remotes")
remotes::install_github("{{ gh_repo }}"){{ packagename }} provides a minimal cohort dataset that can be used to test out the models.
# Load example `cohortdata` included in the package
data("cohortdata")
head(cohortdata, 5)
#> id sex age death_date death_other_causes vaccine_date_1 vaccine_date_2
#> 1 04edf85a M 50 <NA> <NA> <NA> <NA>
#> 2 c5a83f56 M 66 <NA> <NA> <NA> <NA>
#> 3 82991731 M 81 <NA> <NA> <NA> <NA>
#> 4 afbab268 M 74 <NA> <NA> 2021-03-30 2021-05-16
#> 5 3faf2474 M 54 <NA> <NA> 2021-06-01 2021-06-22
#> vaccine_1 vaccine_2
#> 1 <NA> <NA>
#> 2 <NA> <NA>
#> 3 <NA> <NA>
#> 4 BRAND2 BRAND2
#> 5 BRAND1 BRAND1The function make_vaccineff_data allows defining
different aspects of the study design—such as vaccination dates,
immunization delays, and potential confounding factors—and creates an
object of class vaccineff_data. Its output is used to
estimate VE using a Cox model regression invoked by the function
estimate_vaccineff, which returns the object
vaccineff.
The proportional hazard assumption can be tested both through the
Schoenfeld test and visually using the plot method by
setting type = "loglog".
# Create `vaccineff_data`
vaccineff_data <- make_vaccineff_data(
data_set = cohortdata,
outcome_date_col = "death_date",
censoring_date_col = "death_other_causes",
vacc_date_col = "vaccine_date_2",
vaccinated_status = "v",
unvaccinated_status = "u",
immunization_delay = 15,
end_cohort = as.Date("2021-12-31"),
match = TRUE,
exact = "sex",
nearest = c(age = 1)
)
# Estimate VE
ve <- estimate_vaccineff(vaccineff_data, at = 180)
# Print summary of VE
summary(ve)
#> Vaccine Effectiveness at 180 days computed as VE = 1 - HR:
#> VE lower.95 upper.95
#> 0.7109 0.5245 0.8243
#>
#> Schoenfeld test for Proportional Hazards assumption:
#> p-value = 0.0631
# Generate loglog plot to check proportional hazards
plot(ve, type = "loglog")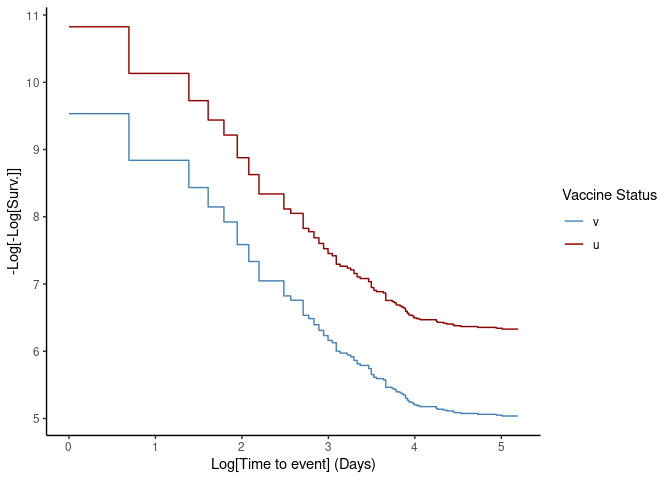
More details on how to use {{ packagename }} can be found in the online documentation as package vignettes, and in the articles “Get Started with vaccineff” and “Introduction to cohort design with vaccineff”.
To report a bug or to request a new feature please open an issue.
Contributions to {{ packagename }} are welcomed. Contributions are welcome via pull requests.
Contributors to the project include:
Authors: David Santiago Quevedo and Zulma M. Cucunubá (maintainer)
Contributors: Geraldine Gómez, Pratik Gupte, Érika J. Cantor, Santiago Loaiza, Jaime A. Pavlich-Mariscal, Hugo Gruson, Chris Hartgerink, Felipe Segundo Abril-Bermúdez, Joshua W. Lambert, Julian Otero
Please note that the {{ packagename }} project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.