

The development version can be installed from GitHub. Installation of
OpeNoise from GitHub is easy using the
devtools package.
# install.packages("devtools")
devtools::install_github("Arpapiemonte/openoise-analysis")This tutorial explains how to use the OpeNoise library. It works on acoustic data acquired with sound level meters instrument. Input dataset format is showed in internal examples that you can access them with the data() function.
library(OpeNoise)
data("PTFA")
head(PTFA)[1:3, 1:6]
#> date LAeq LZFmin.6.3 LZFmin.8.0 LZFmin.10.0 LZFmin.12.5
#> 2 2022-03-07 10:12:16 43.9 31.6 34.7 59.9 74.1
#> 3 2022-03-07 10:12:17 44.6 33.7 43.9 59.6 58.9
#> 4 2022-03-07 10:12:18 44.5 46.0 52.2 51.3 57.2Function calculate energetic average of vector of values in dB. RoundTo function round value at 0.5.
energetic.mean(PTFA$LAeq)
#> [1] 45.7
x <- energetic.mean(PTFA$LAeq)
RoundTo(x, 0.5)
#> [1] 45.5Function calculate energetic average weighted of vector’s values in dB respect to vector’s time like string in format “HH:MM:SS”
energetic_w.mean(c(55.2, 88.6), c("03:22:52", "08:55:33"))
#> [1] 87.2Function return reverse percentile of un vector’s values.
AcuPercentile(PTFA$LAeq)
#> L1 L5 L10 L50 L90 L95 L99
#> 53.747 48.600 47.200 44.400 43.100 43.000 42.700
RoundTo(AcuPercentile(PTFA$LAeq), 0.5)
#> L1 L5 L10 L50 L90 L95 L99
#> 53.5 48.5 47.0 44.5 43.0 43.0 42.5data("exampleHourlyData")
AcuDNPercentile(df = exampleHourlyData,
parameter = "leq",
from = "5",
to = "22",
period = "night")[1:5]
#> [[1]]
#> L1 L5 L10 L50 L90 L95 L99
#> 72.767 72.235 70.940 69.550 51.950 51.420 50.204
#>
#> [[2]]
#> L1 L5 L10 L50 L90 L95 L99
#> 71.448 70.840 70.620 69.850 50.750 48.425 47.285
#>
#> [[3]]
#> L1 L5 L10 L50 L90 L95 L99
#> 71.843 71.615 71.060 69.550 52.250 50.770 48.794
#>
#> [[4]]
#> L1 L5 L10 L50 L90 L95 L99
#> 72.656 72.080 71.280 70.000 52.560 51.350 49.550
#>
#> [[5]]
#> L1 L5 L10 L50 L90 L95 L99
#> 70.770 70.650 70.500 69.600 51.450 49.575 48.795Function return energetic average with hourly aggregation.
HourlyEmean(PTFA, "LAeq", timeZone = "Europe/Rome")
#> date LAeq
#> 1 2022-03-07 10 45.7function retun seconds from hour, minutes and seconds.
hour <- 5
minute <- 25
second <- 50
deco.time(hour, minute, second)
#> [1] "Time decomposition from hours, minutes and seconds to seconds:"
#> [1] 19550This is simple function using Gauss’algorithm to return holiday date in according of Gregorian calendar.
HolidaysDate(2024)
#> [1] "2024-01-01" "2024-01-06" "2024-04-25" "2024-05-01" "2024-06-02"
#> [6] "2024-08-15" "2024-11-01" "2024-03-31" "2024-04-01"Function return energetic average or simple average with aggregation day (06:00/22:00) or night (22:00/06:00).
data("exampleHourlyData")
df_night <- avr.day.night(exampleHourlyData, variable = "leq", period = "night",
stat = "e_mean")
head(df_night, 5)
#> DATA MEAN MIN MAX
#> 1 2020-12-11 56.1 46.9 60.3
#> 2 2020-12-12 54.9 46.5 60.0
#> 3 2020-12-13 56.5 46.3 61.5
#> 4 2020-12-14 56.5 48.4 61.3
#> 5 2020-12-15 56.9 48.6 62.4
df_day <- avr.day.night(exampleHourlyData, variable = "leq", period = "day",
stat = "e_mean")
head(df_day, 5)
#> DATA MEAN MIN MAX
#> 1 2020-12-11 69.9 64.9 72.3
#> 2 2020-12-12 69.4 63.4 72.6
#> 3 2020-12-13 69.0 60.2 72.1
#> 4 2020-12-14 69.6 64.1 73.2
#> 5 2020-12-15 69.7 64.2 72.9This function return energetic average aggregate:
data("exampleHourlyData")
LdenCalculator(dataframe = exampleHourlyData, variable = "leq", type = "daily")
#> # A tibble: 81 × 6
#> date D_acu D E N Lden
#> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 2020-12-11 69.9 70.4 66 NA NA
#> 2 2020-12-12 69.4 69.7 65.6 56.1 68.9
#> 3 2020-12-13 69 69.3 65.4 54.9 68.4
#> 4 2020-12-14 69.6 70 64.9 56.5 69.1
#> 5 2020-12-15 69.7 70.1 65.1 56.5 69.2
#> 6 2020-12-16 70.3 70.7 65.4 56.9 69.7
#> 7 2020-12-17 70.1 70.5 65.9 57.4 69.7
#> 8 2020-12-18 69.6 69.8 66.7 57.2 69.3
#> 9 2020-12-19 69.1 69.4 66.1 56.6 68.9
#> 10 2020-12-20 69.2 69.5 66.5 54.7 68.7
#> # ℹ 71 more rows
LdenCalculator(dataframe = exampleHourlyData, variable = "leq", type = "total")
#> # A tibble: 1 × 4
#> D E N Lden
#> <dbl> <dbl> <dbl> <dbl>
#> 1 69.8 66.3 57.6 69.4Function calculate energetic sum or difference of values
dbsum(x = 55, y = 33, operator = 1)
#> [1] 55.02732dbsum(x = c(55 , 66), y = c(45, 50), operator = 1)
#> [1] 55.41393 66.10774
dbsum(x = c(70 , 68), y = c(55, 66), operator = -1)
#> [1] 69.86045 63.67077Function calculate SEL (single event level)
SELcalc(x = 66.8, t = 938)
#> [1] 96.52203PlotNoiseTimeHistory(df = PTFA, variable = "LAeq", mp = "PTFA", y_lim = c(40, 60))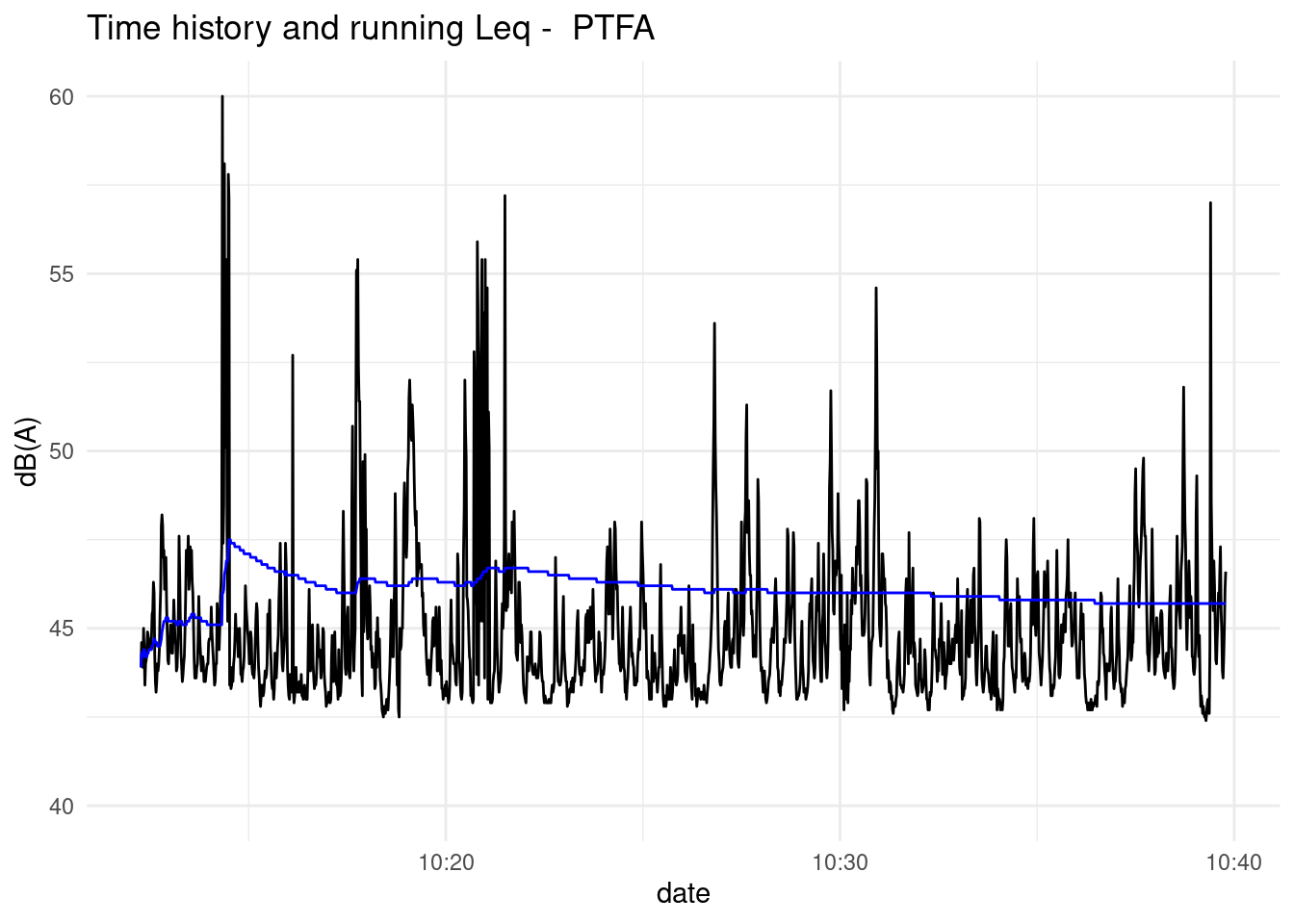
PlotNoiseTHcompare function shows Leq’s time history with frequency components
PlotNoiseTHcompare(df = PTFA,
variable = "LAeq",
listvar = c("LZFmin.100",
"LZFmin.40.0"),
mp = "PTFA",
runleq = FALSE)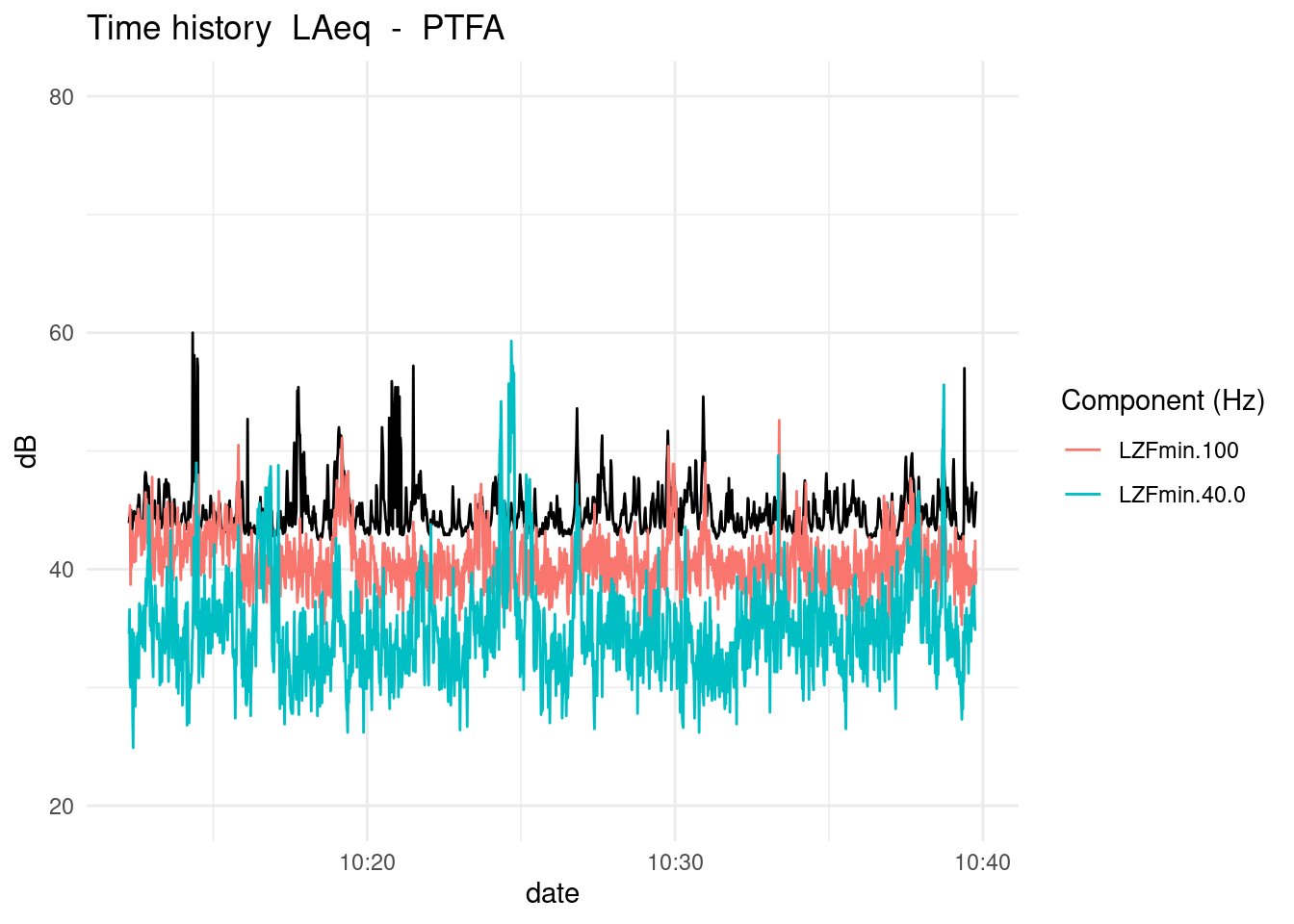
PlotSpectrogram(PTFA, coLs = c(3:38), plot_title = "Spectrogram")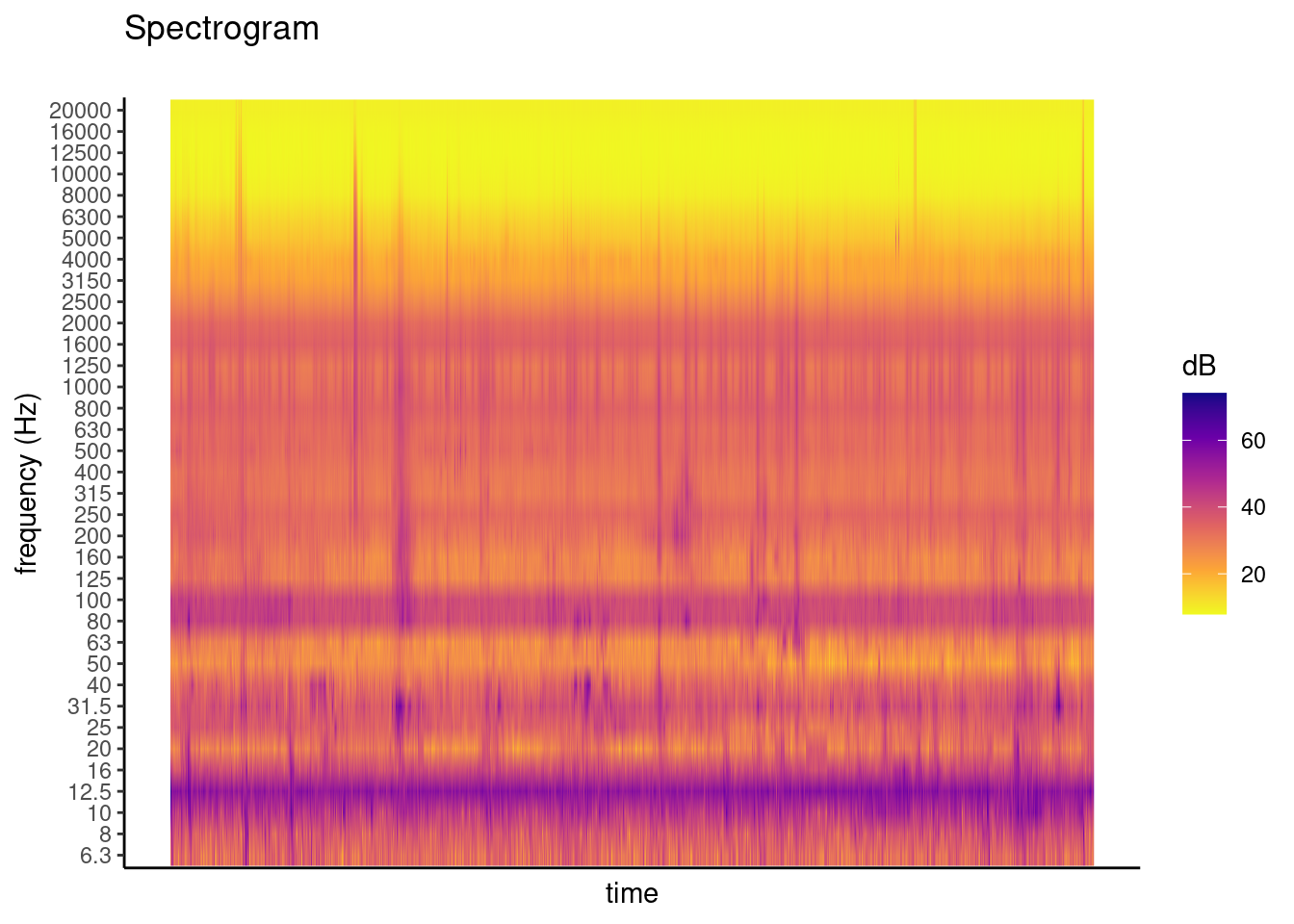
AcousticQuantilePlot function plot acoustic quantile aggregate by hour
library(lubridate)
datasetI <- dataset_impulsive1
datasetH <- dfImpulsiveTrasform(datasetI)
datasetH$date <- ymd_hms(as.character(datasetH$date))
AcousticQuantilePlot(df = datasetH, Cols =c(3:38), Quantile =0.95,
TimeZone = "UTC")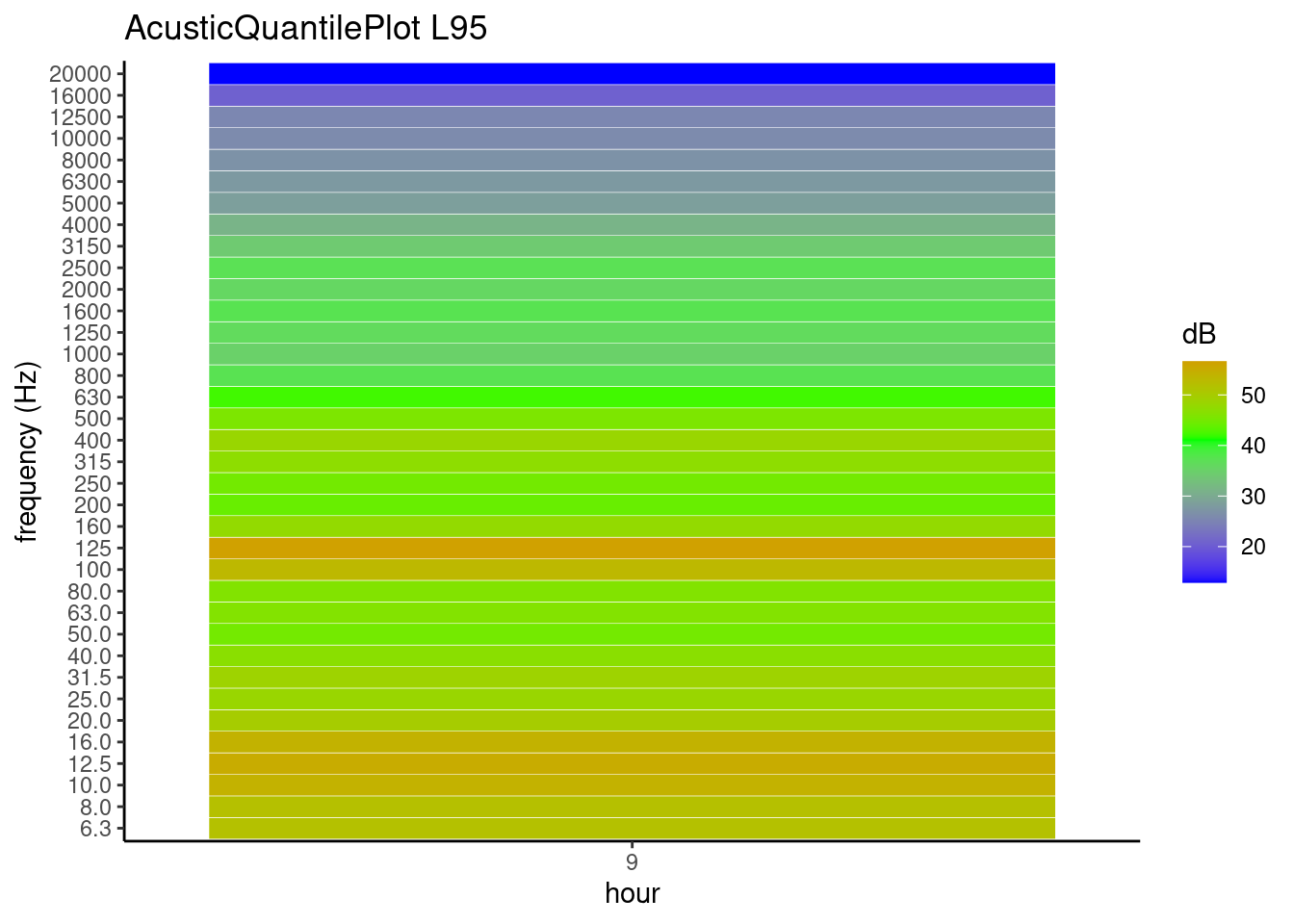
This function search tonal components in acoustic measure in according of Italian law.
search.tone(PTFA[, c(3:38)], statistic = energetic.mean, plot.tone = T)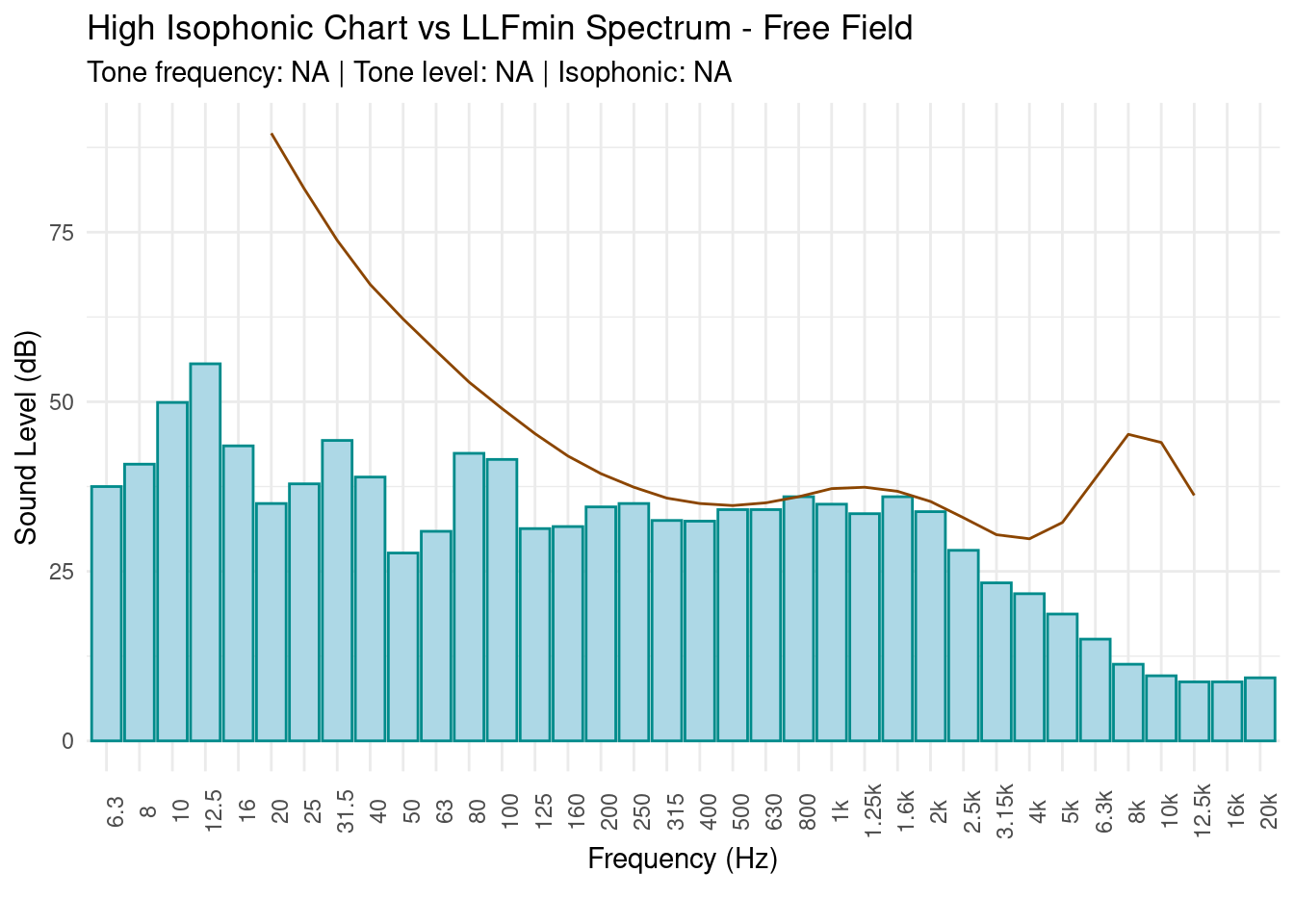
This function search impulsive events in acoustic measure
data("dataset_impulsive2")
results <- searchImpulse(dataset_impulsive2)
results$dfPeaks
#> ymax xmax startPeak stopPeak date cri1 cri2
#> 1 97.2 2795 2794 2818 2022-05-06 14:30:54 y 5
#> 2 96.3 2595 2594 2622 2022-05-06 14:30:34 y 5
#> 3 96.1 1570 1564 1591 2022-05-06 14:28:51 y 4
#> 4 91.1 1978 1977 2000 2022-05-06 14:29:32 y 5
#> 5 91.0 1773 1771 1794 2022-05-06 14:29:11 y 5
#> 6 90.9 2401 2397 2420 2022-05-06 14:30:14 y 4
#> 7 90.2 1159 1158 1183 2022-05-06 14:28:10 y 5
#> 8 90.2 1389 1387 1410 2022-05-06 14:28:33 y 5
#> 9 88.8 2198 2197 2218 2022-05-06 14:29:54 y 4
#> 10 86.7 939 936 959 2022-05-06 14:27:48 y 5results$Plot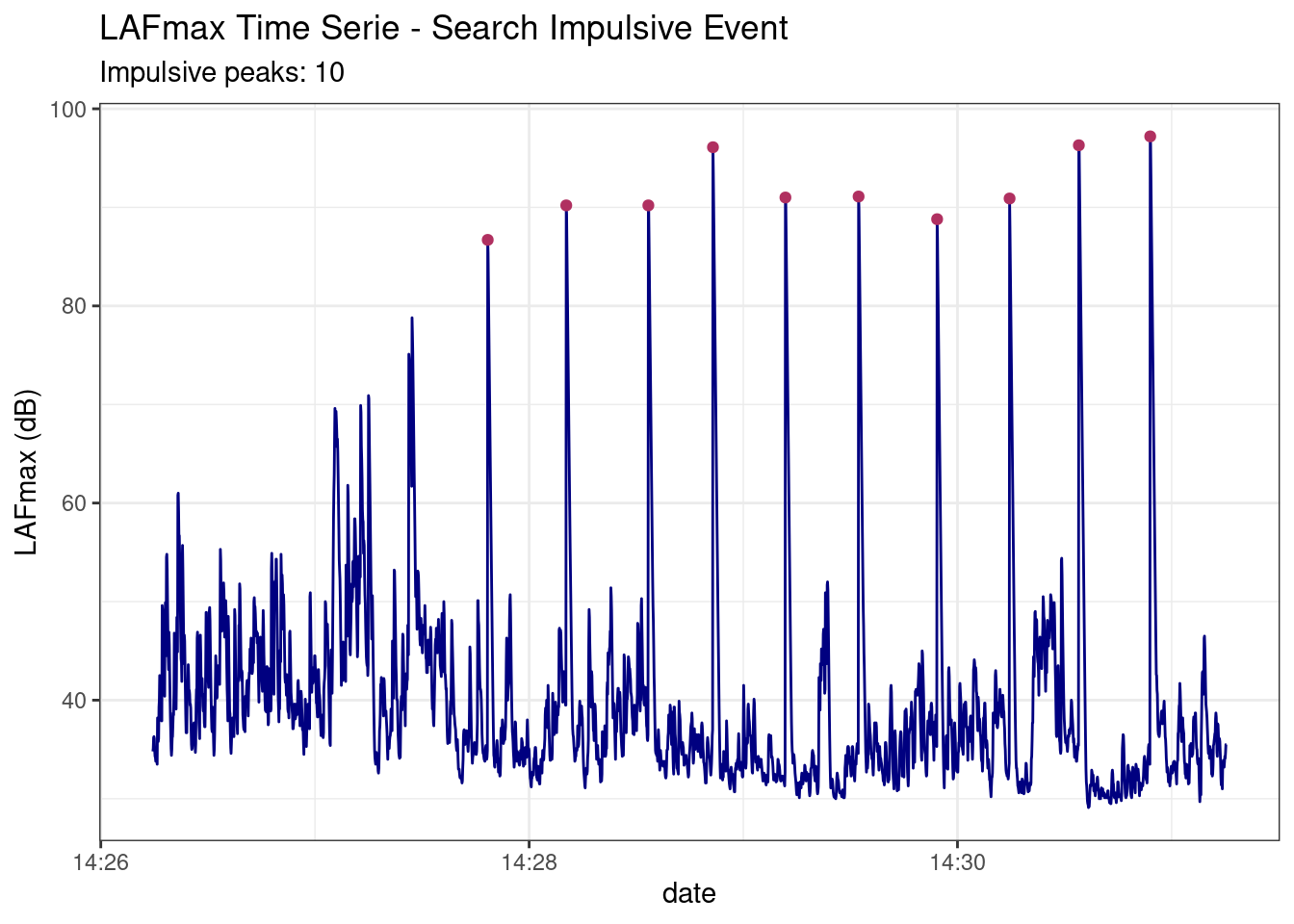
data("dataset_impulsive2")
head(dataset_impulsive2, 3)[, 1:5]
#> date LAeq LASmax LAF LAFmax
#> 2 2022-05-06 14:26:14.600 34.8 40.4 34.5 34.8
#> 3 2022-05-06 14:26:14.700 35.0 40.1 34.6 35.1
#> 4 2022-05-06 14:26:14.800 37.0 39.8 35.7 36.2dfT <- dfImpulsiveTrasform(dfImpulsive = dataset_impulsive2,
statistic = energetic.mean)
head(dfT, 3)[, 1:5]
#> date LAeq LZeq.6.3 LZeq.8.0 LZeq.10.0
#> 1 2022-05-06 14:26:14.6 34.8 29.4 37.5 43.7
#> 2 2022-05-06 14:26:14.7 35.0 31.5 34.9 38.5
#> 3 2022-05-06 14:26:14.8 37.0 28.2 37.1 35.2library(OpeNoise)
library(lubridate)
data("dataset_impulsive1")
data("dfBW")
# dataset handling
df_Imp_sec <- dfImpulsiveTrasform(dataset_impulsive1,
statistic = energetic.mean)
df_Imp_sec$date <- ymd_hms(df_Imp_sec$date, tz = "Europe/Rome")
# extraction of frequency bands from the dataset
freqDF <- df_Imp_sec[, grep("LZeq\\.", names(df_Imp_sec))]
################################################################################
# INTRUSIVENESS INDEX CALCULATION FUNCTION
################################################################################
dfa <- freqDF # Environmental dataset simulation
dfr <- freqDF
# Residual dataset simulation by subtracting 4 from dfa
dfr[c(5,8,12,15), ] <- dfr[c(5,8,12,15), ] - 4
BW <- dfBW$BW # bandwidth
# application of the function
IntrusiveIndex(dfa, dfr, BW)
#> [1] "10 Intrusivity Index is negligible"